mainmenu
- About Center
- Members
- Research
- Publications
- Facilities
- Super-depth Optical Imaging Lab
- Ultrafast Infrared Spectroscopy Lab
- Ultrafast IR-Visible Spectroscopy Lab
- Instrument and Chemistry Lab
- Molecular Imaging Lab
- Multidimensional Infrared Spectroscopy Lab
- Sample Preparation Lab
- Optical Frequency Comb Spectroscopy Lab
- Single Molecule Imaging Lab
- Spectroscopic Imaging Lab
- Theory and Computation
- Open Access Facility
- Collaborative Research Equipment
- Quantum Spectroscopy Lab
- Ultrafast Raman Probe Spectroscopy Lab
- Equipment Inventory
- Board
Posters and Short Descriptions
sHaRPer: algorithm that acquires kinetic information from slow transitions in smFRET measurements
2017 KPS Spring Meeting
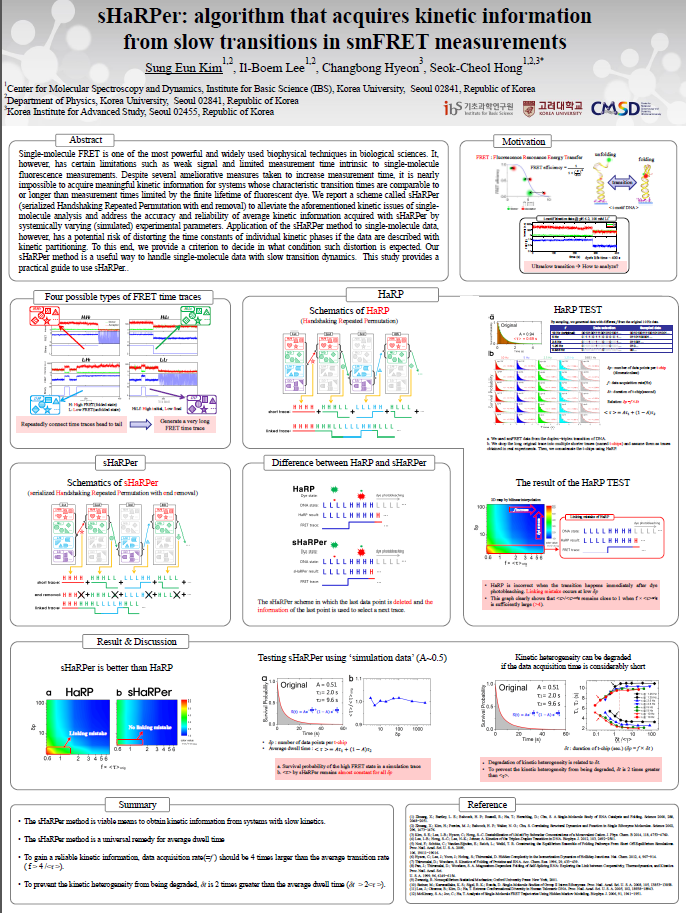
Single-molecule FRET is one of the most powerful and widely used biophysical techniques in biological sciences. It, however, has certain limitations such as weak signal and limited measurement time intrinsic to single-molecule fluorescence measurements. Despite several ameliorative measures taken to increase measurement time, it is nearly impossible to acquire meaningful kinetic information for systems whose characteristic transition times are comparable to or longer than measurement times limited by the finite lifetime of fluorescent dye. We report a scheme called sHaRPer (serialized Handshaking Repeated Permutation with end removal) to alleviate the aforementioned kinetic issues of single-molecule analysis and address the accuracy and reliability of average kinetic information acquired with sHaRPer by systemically varying (simulated) experimental parameters. Application of the sHaRPer method to single-molecule data, however, has a potential risk of distorting the time constants of individual kinetic phases if the data are described with kinetic partitioning. To this end, we provide a criterion to decide in what condition such distortion is expected. Our sHaRPer method is a useful way to handle single-molecule data with slow transition dynamics. This study provides a practical guide to use sHaRPer.